Plant cuticle, which covers the plant surface, consists of waxes and cutins, and is associated with plant drought, cold, and salt re-sistance. Hitherto, at least 47 genes participating in the formation of plant cuticle have been cloned from Arabidopsis thaliana, Oryza sativa, Zea mays, Ricinus communis, Brassica napus, and Medicago truncatula; and about 85% of them encode proteins sharing above 50% identities with their rice homologous sequences. These cloned cuticle genes were mapped in silico on different chromosomes of rice and Arabidopsis, respectively. The mapping results revealed that plant cuticle genes were not evenly distrib-uted in both genomes. About 40% of the mapped cuticle genes were located on chromosome 1 in Arabidopsis, while 20% of the mapped cuticle genes were located on chromosome 2 but none on chromosome 12 in rice. Some cloned plant cuticle genes have several rice homologous sequences, which might be produced by chromosomal segment duplication. The consensus map of cloned plant cuticle genes will provide important clues for the selection of candidate genes in a positional cloning of an unknown cuticle gene in plants.
Forty-seven cuticle-related genes have been cloned in Arabidopsis, rice, maize, Brassica, Medicago, and Ricinus. These genes were in silico mapped in Arabidopsis and rice genomes. The consensus maps of cuticle-related genes will serve as helpful references for cuticle-related gene cloning in plants, especially in important crops such as barley and wheat. The comparison of the rice and Arabidopsis maps revealed that the amount of cloned cuticle genes was similar between Arabidopsis and rice, and most of the correspond-ing genes in both Arabidopsis and rice shared high identity. The micro-synteny analysis of segments carrying homolo-gous genes in rice revealed that some duplicated segments had undergone large-scale changes that could create new functional genes to various environmental stresses.
|
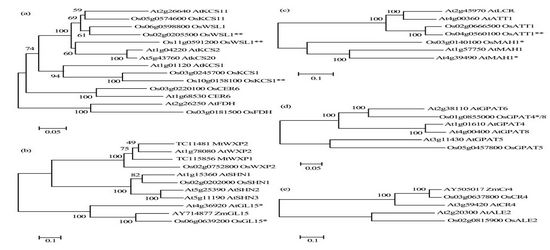
Phylogenetic trees of protein families involved in cuticle formation. The protein members of KCS (a), CYP(b), TF(c), GPAT(d) and receptor kinase are shown. The scale bar of 0.1 represents 10% sequence divergence; bootstrap values are given above the branches
(Picture/Sciences in Cold and Arid Regions) |
|
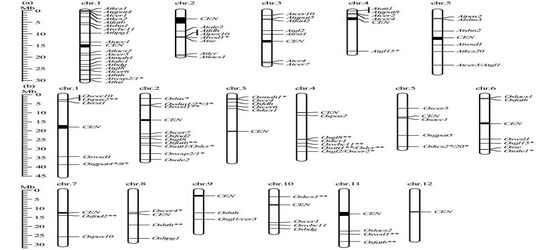
(a) In silico mapping of cloned cuticle genes on Arabidopsis (a) and rice (b) chromosomes. CEN represented centromeric regions
(Picture/Sciences in Cold and Arid Regions) |
|
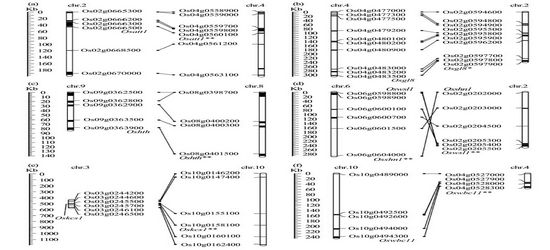
Rice synteny regions carrying homologous sequences of cloned cuticle genes Osatt1(a), Osgl8(b), Oshth(c), Oswsl1(d), Oskcs1(e), and Oswbc11(f). The rice homologous sequences are linked by lines
(Picture/Sciences in Cold and Arid Regions) |







